Structuring the protein sequence space
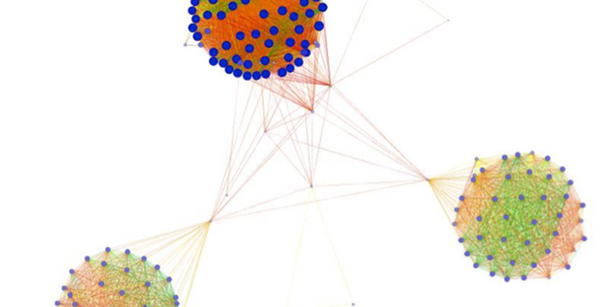
Many biological and evolutionary questions are related to the structure of the protein sequence space and its partitioning into substructures…
☎ +43 1 4277 91280
✉ thomas.rattei(at)univie.ac.at

» Full publications list
» Google Scholar
Thomas Rattei’s work covers a wide spectrum of topics from bioinformatics, genome and metagenome analysis and systems biology. He has long-standing expertise in developing and applying computational methods for the interpretation of large-scale sequence information. The international reputation of his research group triggered their involvement in numerous international (meta-) genome sequencing and analysis consortia.
Thomas’ research activities not only cover individual, project-specific questions but also general problems in bioinformatics, computational infrastructure, and large-scale biological databases. Furthermore, his group develops novel, genome-based computational approaches for studying molecular inter-species interactions, such as between hosts and pathogens, between symbionts, or in microbial ecosystems.
Thomas and his team maintain and develop internationally relevant resources in computational biology, such as the web portals phendb.org, vogdb.org and effectivedb.org for microbial trait prediction, virus orthologous groups and protein families, and bacterial secreted proteins and secretion systems.

Many biological and evolutionary questions are related to the structure of the protein sequence space and its partitioning into substructures…
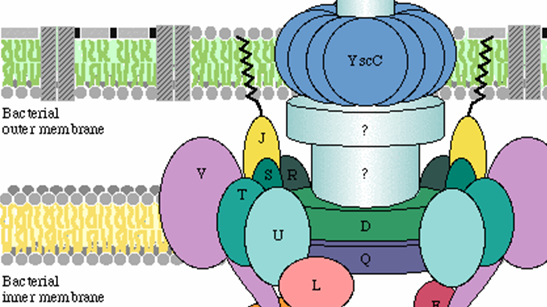
Annoying for most of us, but dangerous or even life threatening for particular persons: infections by pathogenic bacteria are never welcome.

Although microbial communities (“microbiomes”) are central to life on earth, they largely remained terra incognita until recently. By developing a…
Technician
PhD Student
Postdoc
Data Reuse Consortium, Wagner M, Rattei T, Osvatic J, Speth D. A roadmap for equitable reuse of public microbiome data. Nature Microbiology. 2025 Sept 26;10(10):2384-2395. Epub 2025 Sept 26. doi: 10.1038/s41564-025-02116-2
Tocino-Márquez I, Zehl M, Séneca J, Pjevac P, Felkl M, Becker CFW et al. The bacterial community of the freshwater bryozoan Cristatella Mucedo and its secondary metabolites production potential. Scientific Reports. 2025 Aug 26;15(1):31456. doi: 10.1038/s41598-025-17084-0
Gelabert P, Oberreiter V, Straus LG, Morales MRG, Sawyer S, Marín-Arroyo AB et al. Author Correction: A sedimentary ancient DNA perspective on human and carnivore persistence through the Late Pleistocene in El Mirón Cave, Spain. Nature Communications. 2025 Jan 17;16(1):779. 779. doi: 10.1038/s41467-025-56198-x
Gelabert P, Oberreiter V, Straus LG, Morales MRG, Sawyer S, Marín-Arroyo AB et al. A sedimentary ancient DNA perspective on human and carnivore persistence through the Late Pleistocene in El Mirón Cave, Spain. Nature Communications. 2025 Jan 2;16(1):107. doi: 10.1038/s41467-024-55740-7
Hämmerle M, Guellil M, Trgovec-Greif L, Cheronet O, Sawyer S, Ruiz-Gartzia I et al. Screening great ape museum specimens for DNA viruses. Scientific Reports. 2024 Nov 30;14(1):29806. doi: 10.1038/s41598-024-80780-w
Thomas Rattei is part of the FWF-funded Cluster of Excellence (CoE)

